Bub lab
p: 514 398 8148
Our aim is to understand the dynamics of excitable cell networks found in the heart and brain. A big part of the research program is to develop imaging and computational methods - in particular new microscopy modalities, analysis and simulation programs, optogenetic techniques, and new ultra-fast sensors - and use these tools to study complex cardiac and neuronal excitable systems. Research themes goes into detail on new imaging techniques ( instrumentation), the data we collect in cardiac tissues ( excitable systems), and how we can control cells in real-time ( optogenetics).
research themes
The lab works on several closely interrelated research themes. If you are interested in being involved as either undergraduate trainee, graduate student, or postdoc, please contact Dr Gil Bub (gil.bub@mcgill.ca).instrumentation
Excitable cells in the heart and brain generate fast signals (action potentials) that have to be captured using specialized detectors at over a thousand frames per second. At the same time, the structure of these cells is complex, requiring high resolution detectors and advanced microscopy techniques. We're working on many technologies to help with these issues: three examples - temporal pixel multiplexing, RAP imaging, and remote focusing microscopy - are shown below.
Temporal pixel multiplexing (TPM) is an imaging method that embeds high speed information into still image frames. Camera chips have millions of pixels which integrate charge generated when light hits their surface over a set integration time. TPM controls the way individual pixels are exposed during a single frame to embed high speed information in static images. This allows a lower resolution high speed movie to be extracted, giving a high-res still and a high-speed movie in one shot.

The TPM prototype used a micro-mirror array (DMD) to selectively expose pixels. The blur in the high res image can be resolved into a moving ball after decoding. The queen isn't moving so she's captured at high resolution.
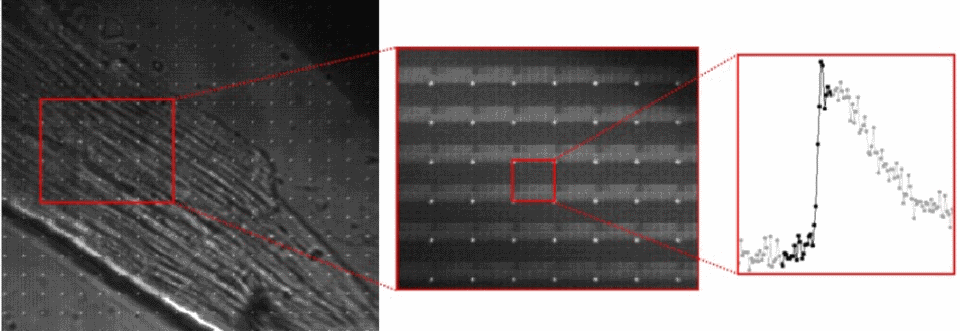
A cardiac cell is loaded with a calcium sensitive dye. A high resolution image is captured with the TPM prototype, which allows the pixel data to be re-arranged to show changes in cell calcium during an action potential.
The original TPM prototype was built using a micromirror array (similar to those used in many consumer overhead projectors) to control pixel exposure times in a off-the-shelf camera. We are now developing CMOS sensors that incorporate TPM technology at the pixel level. Unlike other imaging chips which store data every frame, TPM doesn't move data (or transfer charge) during acquisition allowing it to be extremely fast. Our new chips should be able to image activity at over a million frames/second.
Random Access Parallel (RAP) microscopy (previously called solid-state high throughput (SSHTs) imaging) is a new high-throughput brightfield imaging modality that uses an inverted Newtonian telescope architecture with a high speed LED array to image living samples. RAPs advantage comes from being able to change its field of view without any moving parts or robotics. This allows it to switch views so rapidly that you can effectivly image different, spatially separated samples in parallel.

First (left) and second (right) generation RAP prototypes built around a single parabolic reflector, a 500 fps machine vision camera, and high-speed LED array. The systems both use easily sourced parts and 3D printed components. A parts-list is available in our eLife paper.
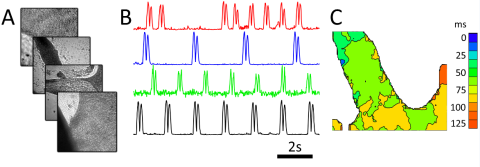
Cardiac monolayer imaging using the first generation prototype. Four cardiac monolayers are imaged simultaneously by interlaving samples. Motion transients show different beating patterns in each monolayer.
The RAP microscope is optimized for capturing dynamic data from model organisms or macroscopic tissues, such as C. elegans travel and wave propagation in cultured cardiac monolayers. The first RAP prototype, built by three undergraduates in the McGill lab, was able to image four samples simultaneously. Our second generation prototype increased the sample count to 80, and we are currently working on new versions with a range of capabilities.
Remote focusing is a microscopy technique, pioneered by Prof. Tony Wilson's Scanning Optical Microscopy group in Oxford, which allows tissue to be scanned in depth without moving the sample or the imaging objective. When combined with laser scanning two-photon microscopy, the technique allows capture of cell structure and function in living tissue.
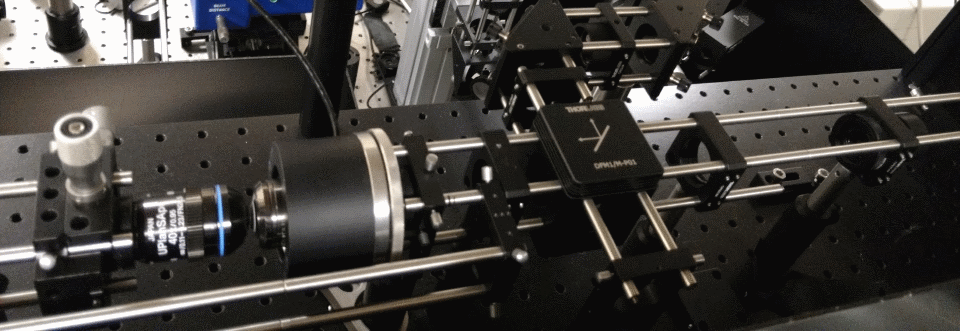
Two-photon microscopy uses infrared lasers to image cells in living tissue. Many labs build their own setups allowing them to test new techniques. This is a high-speed remote focusing microscope currently under development.
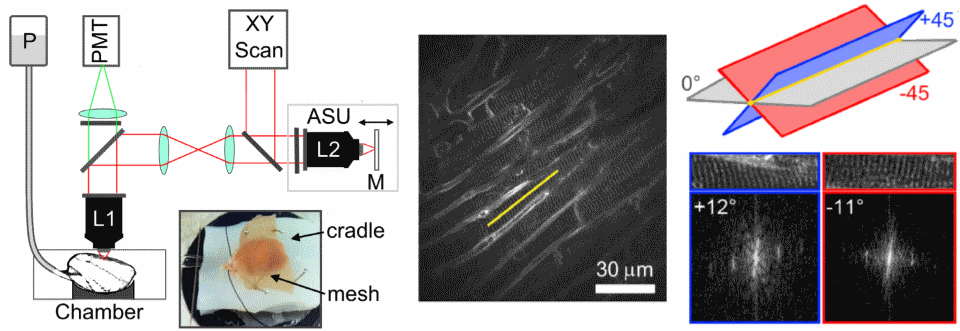
Remote focusing can be used to measure cell structure in the heart. It does this by measuring two oblique images at 45 degrees to the image plane, and using the measured sarcomere angle to calculate the cell's orientation.
One of the main limitations of scanning microscopes is that they're slow as the laser has to visit each point in an image. We're developing new remote focusing microscopes which split the laser into many beamlets, so we can increase acquisition speed. The aim of this project is to be able to measure cell-cell communication in intact heart and brain tissues.
|
more TPM: see the Nature Methods paper and articles in New Scientist & IO9 magazines. more RAP: see the eLife paper. Coming soon - RAP imaging and construction pages. more remote focusing: see Tony Wilson's group original PNAS paper and our Circ Res paper. collaborators: Ed Mann, Bei Li, Alex Corbett , Leonardo Sacconi , Martin Booth, Tony Wilson industry & licencing: Cordin Scientific Imaging, STFC, Oxford University Innovation. |
excitable systems
Excitable media are systems which have the ability to propagate signals spatially without damping. In the heart, waves of excitation synchronise cardiac muscle contraction with every heart beat, and changes in the spatial patterns of these waves cause potentially deadly arrhythmias. We study these waves using mathematical models, intact heart muscle and bioengineered sheets of tissue, using a variety of experimental techniques.
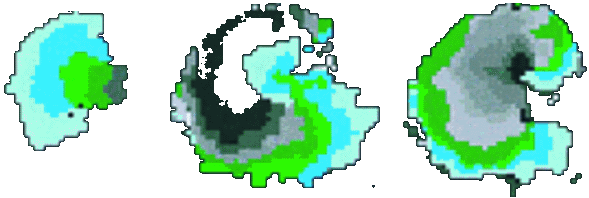
|
|
Isochronal map of spiral wave initiation in a cardiac monolayer. PNAS, 1998. |
Simulations: Target and spiral waves are seen in many physical excitable media due to the relatively simple rules that underpin the behaviour of connected cells, even though the physical processes that drive each system can be extremely complex. These rules can be coded into a simple discrete computer model called a cellular automata
|
for more: the original excitable media pages, optical mapping database, the PNAS and PRL papers.
new software! Jakub Tomek's Ccoffinn Toolkit: paper and download: collaborators: Leon Glass, Alvin Shrier , Emilia Entcheva , Kevin Webb, Rebecca Burton, Matt Daniels. |
optogenetics
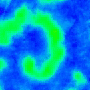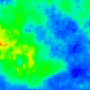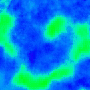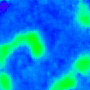
|
|
Nature Photonics, 2015. |
The term optogenetics refers to a set of methods for controlling and imaging genetically modified living tissue with light. Optogenetics has revolutionised the neurosciences and now is becoming increasingly popular in other tissues. In close collaboration with Emilia Entcheva's COOL lab , we've been sensitizing cardiac monolayers to light and seeing how we can control their behaviour.
One goal is to use optogenetics to control macroscopic wave dynamics. Macroscopic excitation waves are found in chemical reactions, slime mold colonies, as well as cardiac and neuronal tissues. In cardiac tissue, the patterns formed by these waves are different in healthy and diseased states. The ability to control wave shape, direction and velocity in cardiac tissue would expose new research targets: for example, since conduction velocity variability often causes re-entrant waves, control of wave velocity allows us to test hypotheses on spiral wave formation in a precise way.
The animated image (top right) shows an example of chirality control (chirality refers to the spirals rotation direction). A spiral wave in a cardiac monolayer (green) is coaxed into rotating in the opposite direction by a projected (red) spiral. Chirality control allows us to observe how spirals respond to stimuli: in this case the new spiral meanders to a prefered location, suggesting that the spiral probes its environment.
Future projects include controlling sympathetic neurons in intact heart and co-cultures, and revisiting the many classical experiments done on light sensitive chemical systems.
|
for more: the Nature Photonics paper and a review in the Journal of Physiology.
collaborators: Emilia Entcheva , Rebecca Burton, Elis Cooper, Leonardo Sacconi , and Alex Corbett . |
selected papers
Ashraf et al., 2021. Random access parallel microscopy. Elife.Biasci et al., 2020. Universal mechanisms for self-termination of rapid cardiac rhythm. Chaos.
Burton et al., 2020. Optical interrogation of sympathetic neuronal effects on macroscopic cardiomyocyte network dynamics. Iscience.
Sacconi et al., 2020. KHz-rate volumetric voltage imaging of the whole zebrafish heart. biorXiv.
Burton et al., 2015. Optical control of excitation waves in cardiac tissue. Nature Photonics.
Botcherby et al., 2013. Fast measurement of sarcomere length ... using remote focusing microscopy. Circulation Research.
Bub et al., 2010. Temporal pixel multiplexing for simultaneous high-speed, high-resolution imaging. Nature Methods.
Bub, Shrier and Glass, 2002. Spiral wave generation in heterogeneous excitable media. Physical Review Letters.
Bub et al., 1998. Bursting calcium rotors in cultured cardiac myocyte monolayers. Proceedings of the National Academy of Sciences.
classes and teaching
Fall term: Mathematical Models in Biology (BIOL 309).A simple CCD simulator for Claire Brown's MLMC 2017 Microscopy Fundamentals course:
Implementations of a 1D CA and Conways Game of Life for Bio 309.
A HTML5 logistic map iterator, a webGL parameter space diagram of the sine map, and webGL cellular automata for Bio 309.
Some more rough demos for Bio 309...
A predator prey agent based CA, a Glass Pattern generator, a mutual inhibition and Fitzhugh Nagumo phase space demo.
The Chaos game, a IFS Barnsely fern demo, a DLA sim, and a Mandelbrot set generator that shows a link to the logistic map.
Many of these have no instructions as they are used by me in class - let me know if you would like to see more details.
people
|
current members: Gil Bub (PI) associate professor Physiology Department McGill University |

| |
|
Thomas Bury Postdoctoral researcher Physiology Department McGill University also working with Leon Glass and Alvin Shrier web page |

|
|
|
Miguel Romero Sepúlveda PhD student Physiology Department McGill University |

|
|
|
Khady Diagne PhD Student Quantitative Life Sciences McGill University web page |

|
|
|
Kiamehr Rahemipour Masters Student Physiology Department McGill University |

|
|
|
Younes Valibeigi Masters Student Physiology Department McGill University |

|
|
|
Karissa Becknel Masters Student Physiology Department McGill University |

|
|
|
Megha Kodancha Undergraduate trainee Physiology Department McGill University |

|
|
|
Benjamin Rudski Undergraduate trainee Physiology Department McGill University |

|
|
|
Glisant Plasa Mackey Glass Summer Research Fellow Physiology Department McGill University |

|
|
|
Leslie Unger Undergraduate trainee Physiology Department McGill University |

|
|
|
Jake Zao Undergraduate trainee Physiology Department McGill University |

|
|
|
prior grad students: | ||
|
Adrienne Caldwell MSc. - graduated 2019 Physiology, McGill co-supervised with Alvin Shrier |
Jakub Tomek DPhil - graduated 2017 DPAG, Oxford web page |
|
|
prior undergrad trainees: Mishal Ashraf (now MSc, UBC), Anthony Tam (now MUHC, McGill), Byuri Sim (now MSc, Shanghai), Yujing Zao (now MsC, McGill Medical Physics), David Gao (now med school, U. Ottawa), Maharshee Karia, Valentine Habrard (MSc, ISAE-SUPAERO, France), Marc Andre Rousseau (now MILA), Judy Chen (now MA Educational Leadership, McGill), Edward Tu, Ila Goshal (MSc, McGill's Hayer lab), Chenhui Zhao (now MSc, McGill), Olivier Nahman Levesque, Vincent Mackay... | ||
